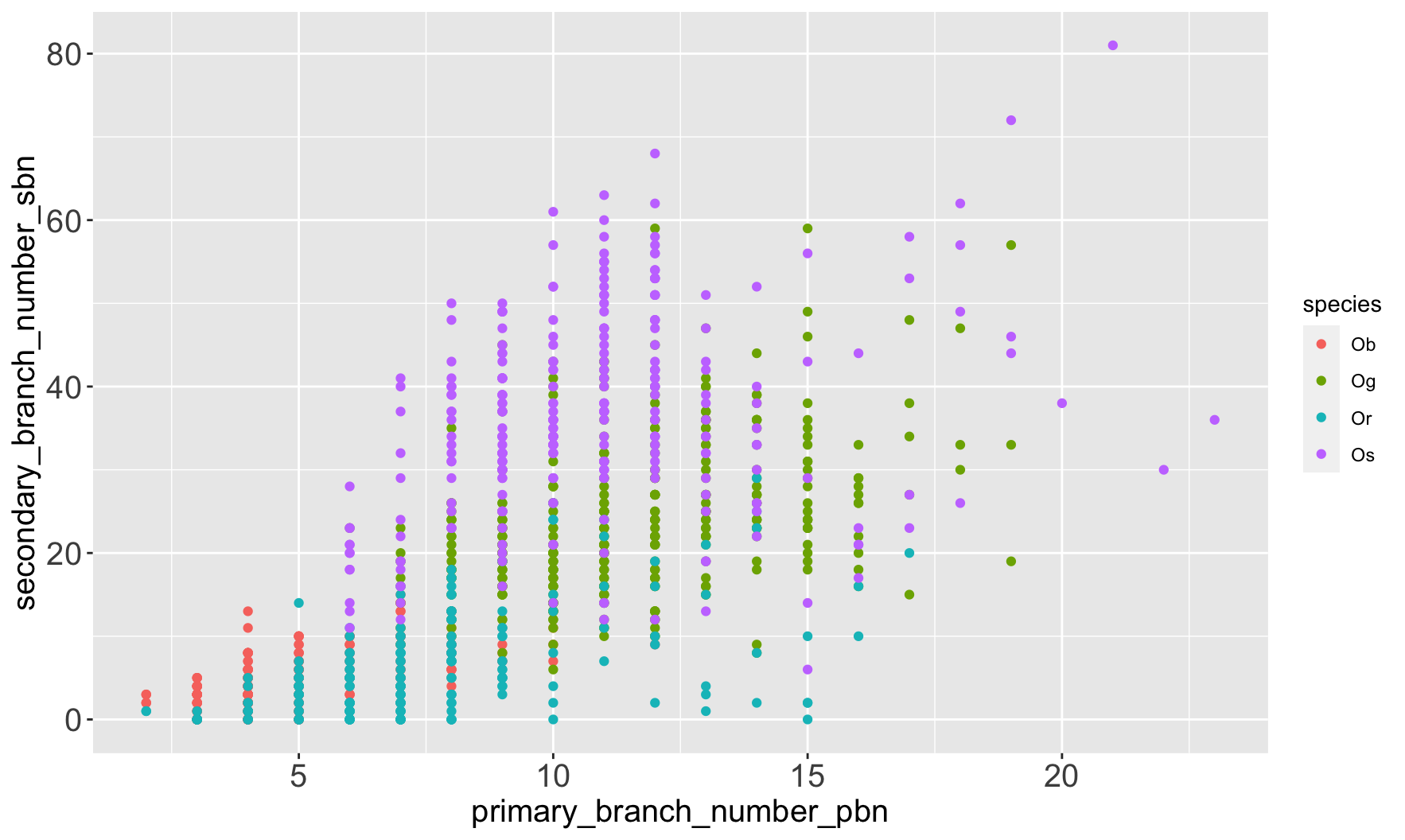
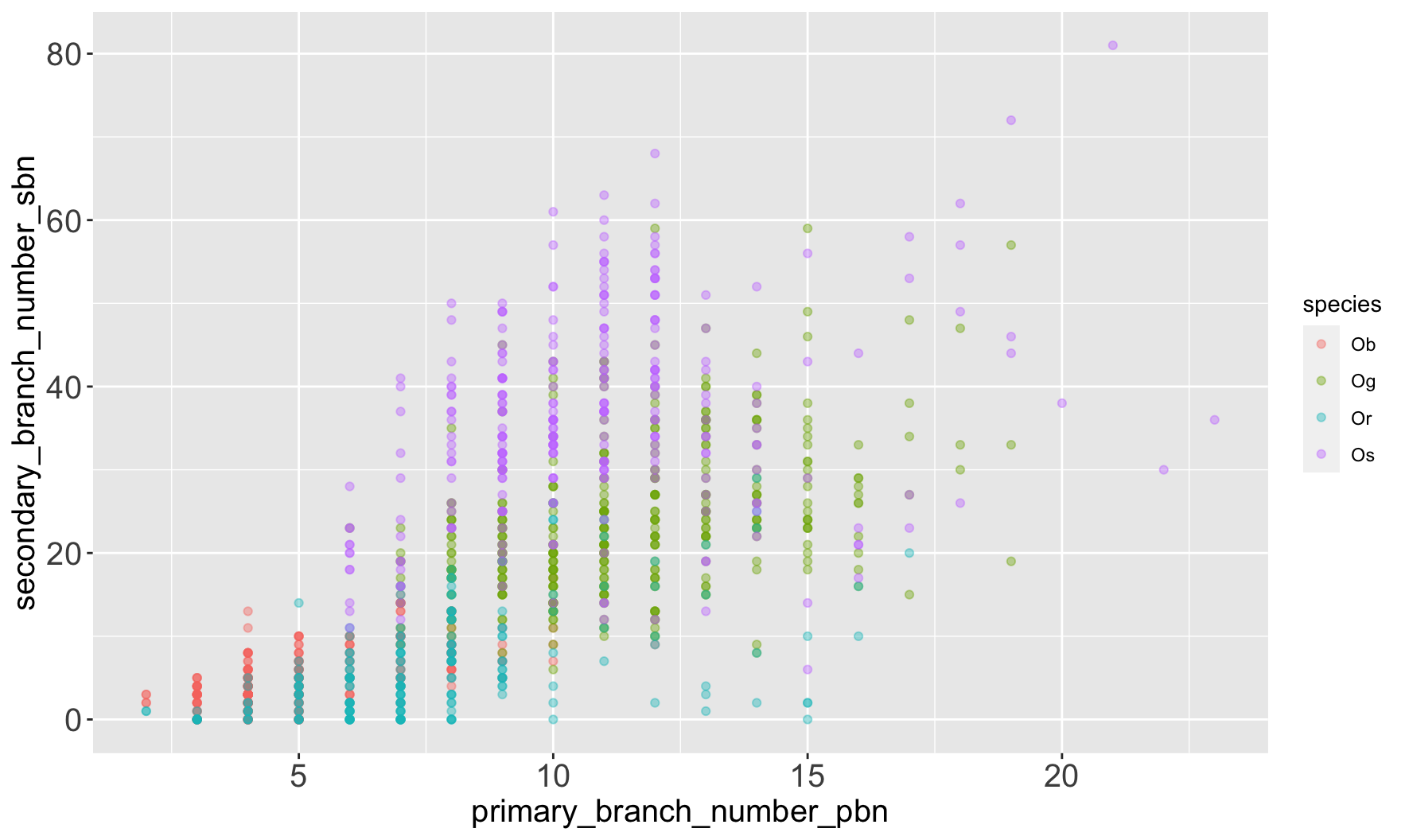
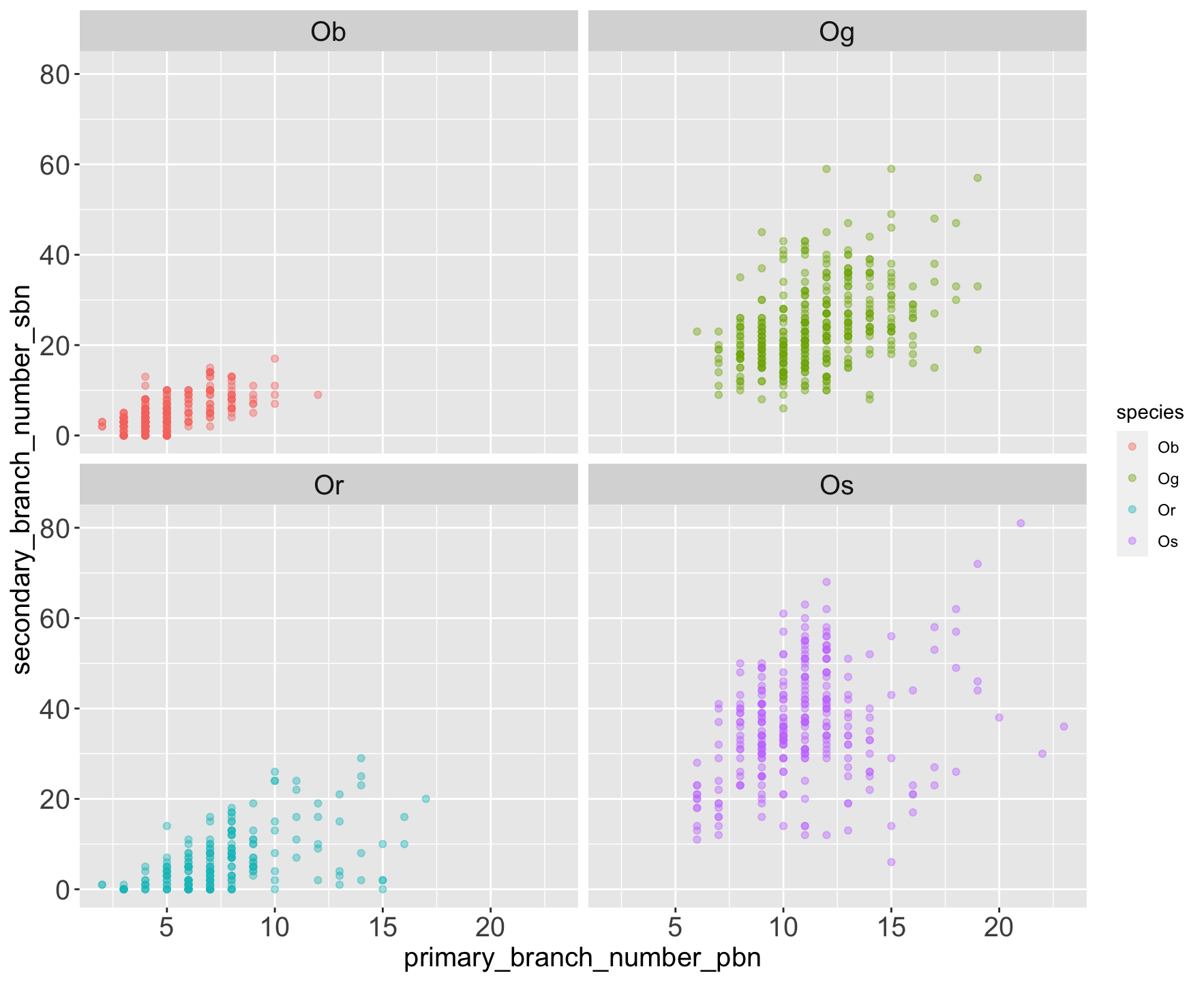
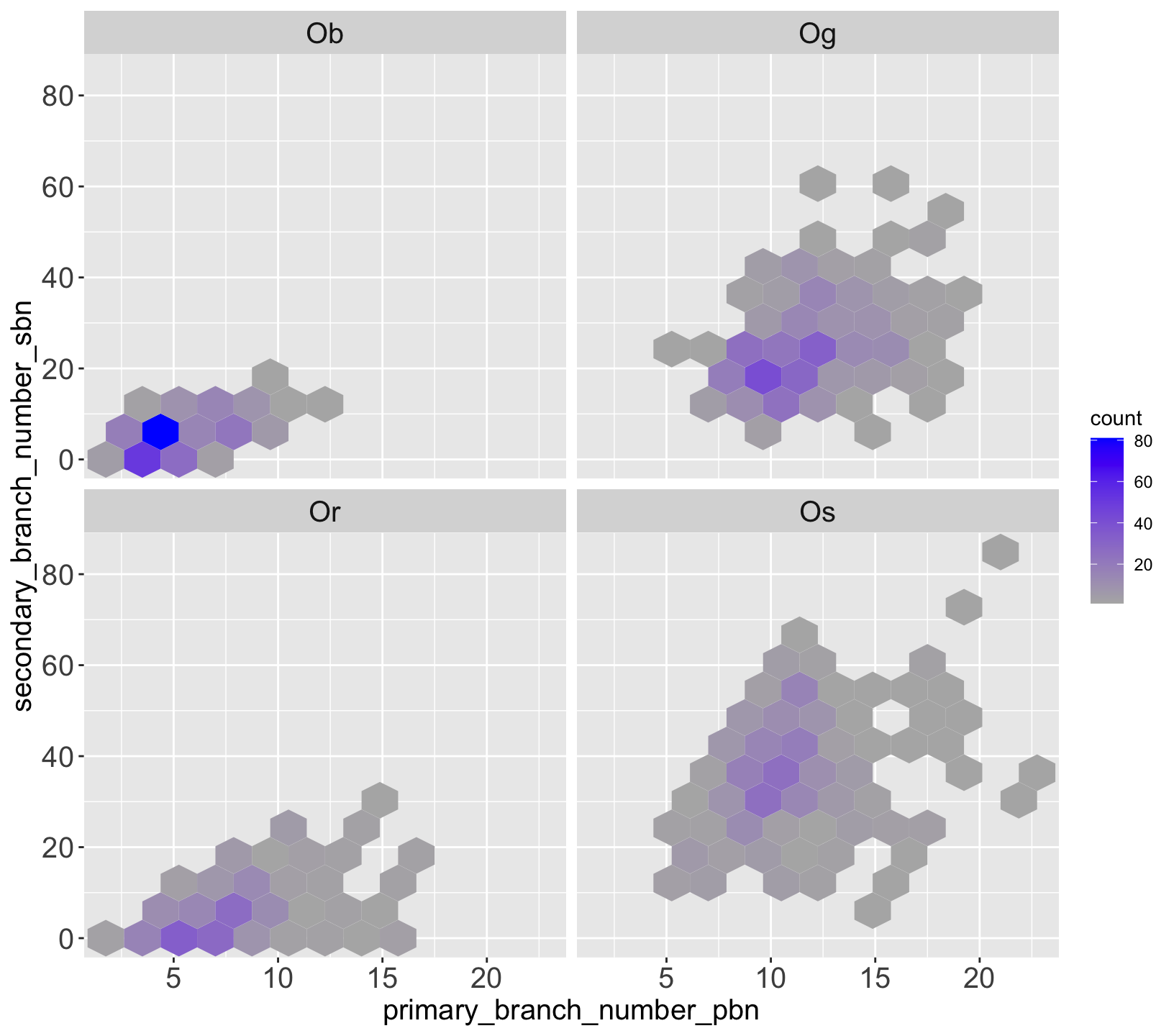
| species_id | species | continent | status |
|---|---|---|---|
| Ob | Oryza barthii | African | Wild |
| Og | Oryza glaberrima | African | Cultivated |
| Or | Oryza rufipogon | Asian | Wild |
| Os | Oryza sativa | Asian | Cultivated |
Explorative Data Analysis
Tidyverse - Part 5
Putting it All Together
The steps of EDA
We have covered almost all steps of what is called Explorative Data Analysis (EDA).
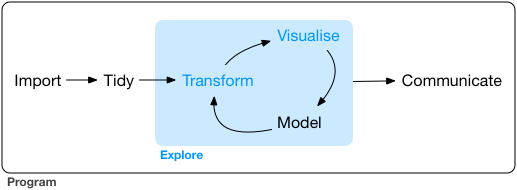
Image Source: R4DS
The Steps of EDA
The explore part is iterative.
- Transform
- Visualize
- Model
A imperative for EDA
To get insights from data that you haven’t faced before,
- Look what’s inside.
- Learn their structure
- Formulate questions
- Try to anwser them with statistical methods.
Briefly, explore them.
Practice Dataset
Transcriptome and Phenotype of multiple Rice species
A set of AP2-like genes is associated with inflorescence branching and architecture in domesticated rice.

- Paper
- DOWNLOAD SUPPLEMENTARY TABLE S3 - 96Kb !It’s a CSV file!
4 rice accessions
Let’s read the Data
Rows: 1,140
Columns: 18
$ id <chr> "Ob01", "Ob01",…
$ species <chr> "Ob", "Ob", "Ob…
$ accession_name <chr> "B197", "B197",…
$ origine_continent <chr> "Africa", "Afri…
$ type_wild_cultivated <chr> "Wild", "Wild",…
$ sowing_localisation <chr> "Cali-Colombia"…
$ replicate_nb_1_2 <dbl> 1, 1, 1, 1, 1, …
$ plant_nb_1_to_3 <dbl> 1, 1, 1, 2, 2, …
$ panicle_nb_1_to_3 <dbl> 1, 2, 3, 1, 2, …
$ rachis_length_rl_in_cm <dbl> 8.10, 6.51, 4.6…
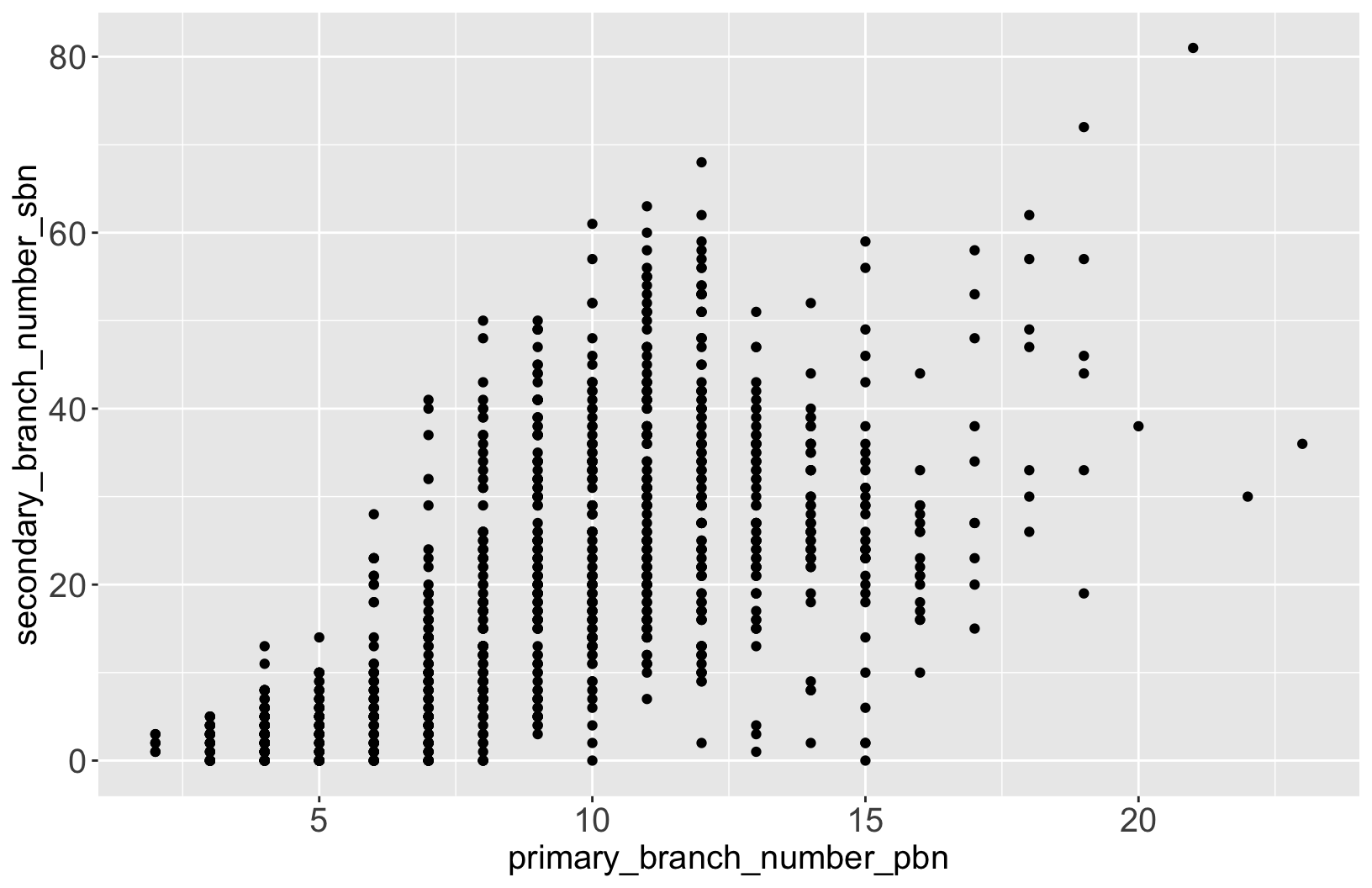
$ primary_branch_number_pbn <dbl> 5, 5, 4, 4, 4, …
$ average_of_primary_branch_length_in_cm_pbl <dbl> 6.73, 6.26, 6.9…
$ average_of_internode_along_primary_branch_in_cm_pbil <dbl> 2.03, 1.63, 1.5…
$ secondary_branch_number_sbn <dbl> 5, 3, 2, 5, 1, …
$ average_of_secondary_branch_length_in_cm_sbl <dbl> 1.74, 1.51, 1.9…
$ average_of_internode_along_secondary_branch_in_cm_sbil <dbl> 0.99, 0.72, 1.6…
$ tertiary_branch_number_tbn <dbl> 0, 0, 0, 0, 0, …
$ spikelet_number_sp_n <dbl> 38, 38, 30, 35,…Goal of the paper
Explore the phenotypic and transcripts of panicle development.
Traits of agronomic interest in the panicle:
- Number of Spikelet,
- Number of Primary branches
- Number of Seconday branches
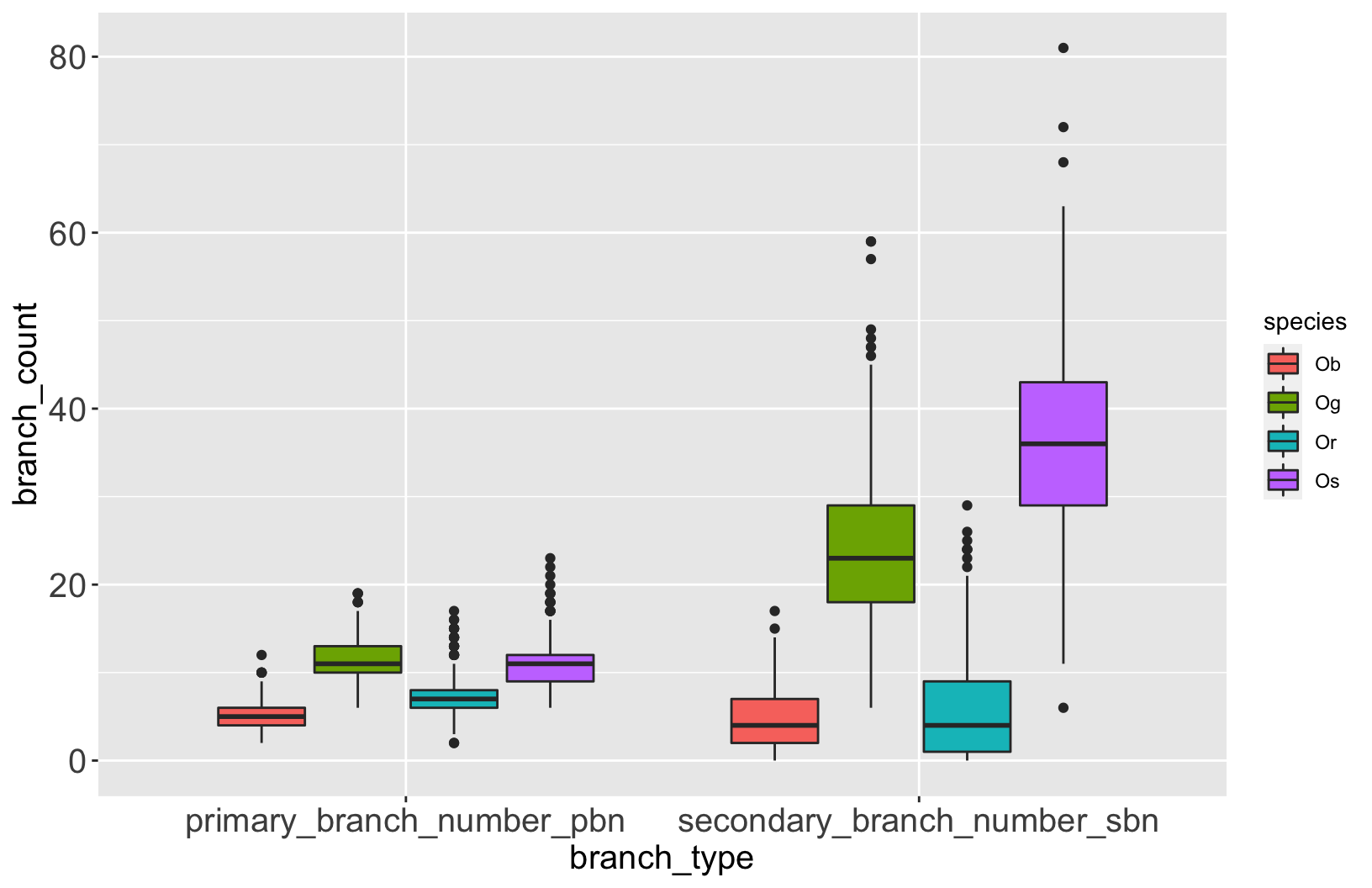
Let’s explore the data
# define colors
rice_colors <-
c(Or = '#b5d4e9',
Os = '#1f74b4',
Ob = '#c0d787',
Og = '#349a37')
# plot
rice %>%
select(species,
primary_branch_number_pbn,
secondary_branch_number_sbn) %>%
pivot_longer(-species,
names_to = 'branch_type',
values_to = 'branch_count') %>%
ggplot(aes(x = branch_type,
y = branch_count,
fill = species)) +
geom_boxplot() +
scale_fill_manual(values = rice_colors)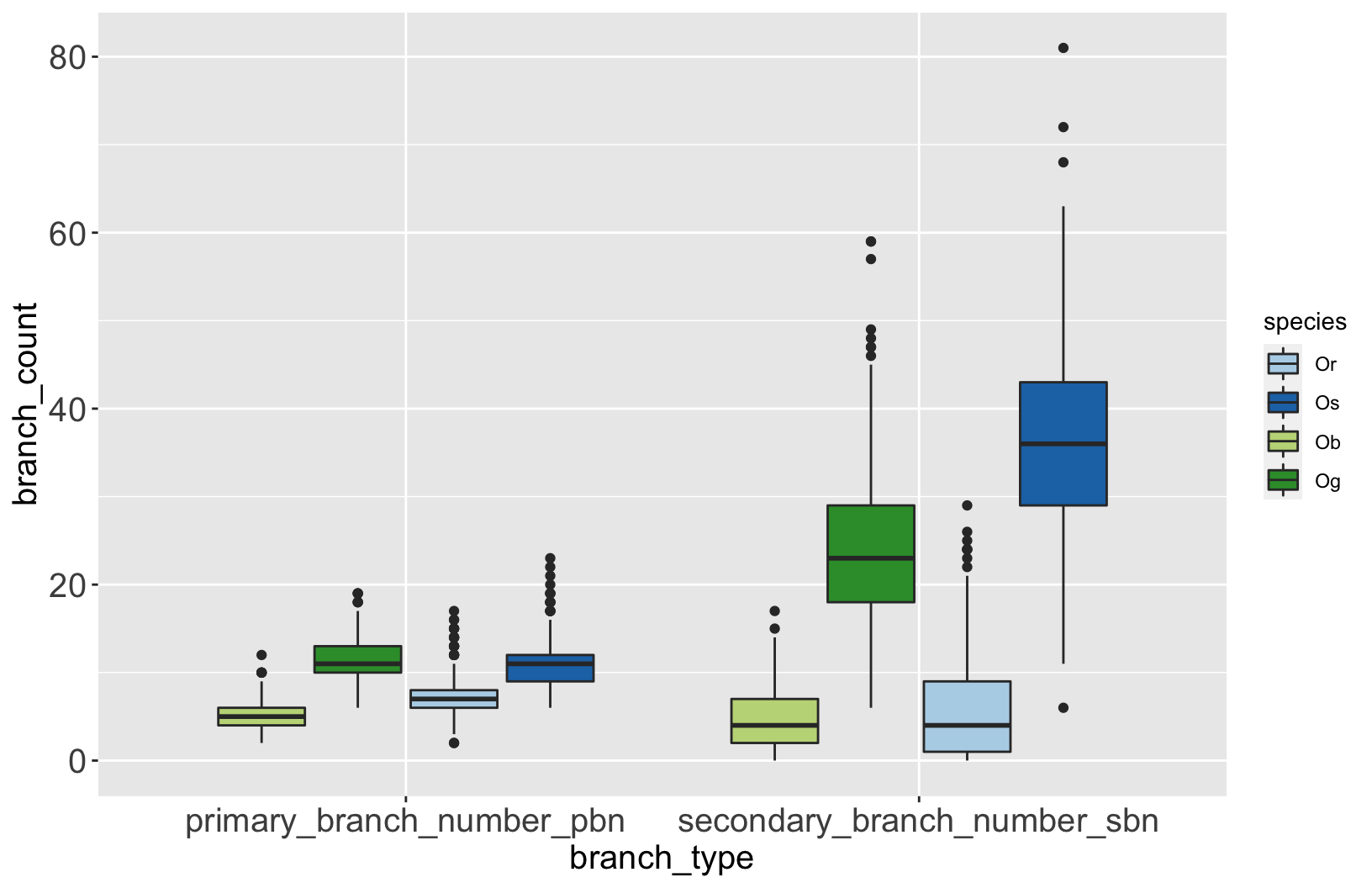
Exercise
Explore the rice panicle dataset.
How are the measured variable distributed? Do they correlate with each other?
Can you find some of the traits that mark the differences between wild and domesticated varieties? And what about the differences between the Asian and African varieties.
Did you manage to anwser all your questions with summary statistics and graphics? Note down questions that are still open and that you think should be tackled with more advanced statistics.