Clean and Tidy Data
Tidyverse - Part 4
Tidy Data
Which Dataset is Tidy?
A common practical way to structure (empirical) data.
- Every column is a variable.
- Every row is an observation.
- Every cell is a single value.
- (Every observational unit is in its own table).
Plus: fixed variables should come first, followed by measured variables.
Reference: An Introduction to Tidy Data
Which Dataset is Tidy?

Source: R4DS - Tidy Data
Semantics of (tidy) Data
Always quoting the Tidy Data article:
- A dataset is a collection of values.
- Every value belongs to a variable and an observation.
- A variable contains all values that measure the same underlying attribute (like height, temperature, duration) across units.
- An observation contains all values measured on the same unit (like a person, or a day, or a race) across attributes
Tools: Tidy data with Tidyr
Tidyr provides functions for:
- Pivoting data.
- Rectangling data.
- Nesting data.
- Combining and splitting columns.
- Make missing values explicit.
Let’s tidy the data from Pangaea
Pangea Data
Remember the dataset from Pangaea?
Rows: 128
Columns: 19
$ Event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL…
$ Latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617…
$ Longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117…
$ `Elevation [m]` <dbl> -339, -339, -339, -339, -339, -339, -339, -…
$ `Sample label (barite-Sr)` <chr> "SL73-1", "SL73-2", "SL73-3", "SL73-4", "SL…
$ `Samp type` <chr> "non-S1", "non-S1", "non-S1", "S1b", "S1b",…
$ `Depth [m]` <dbl> 0.0045, 0.0465, 0.0665, 0.1215, 0.1765, 0.1…
$ `Age [ka BP]` <dbl> 1.66, 3.13, 4.13, 5.75, 7.30, 7.78, 8.65, 9…
$ `CaCO3 [%]` <dbl> 61.5, 55.1, 53.0, 43.4, 41.8, 42.3, 42.6, 3…
$ `Ba [µg/g] (Leachate)` <dbl> 72.6, 64.3, 37.4, 63.5, 101.0, 141.0, 75.2,…
$ `Sr [µg/g] (Leachate)` <dbl> 767, 681, 690, 552, 527, 528, 551, 482, 391…
$ `Ca [µg/g] (Leachate)` <dbl> 188951.9, 163260.4, 162188.7, 125937.6, 124…
$ `Al [µg/g] (Leachate)` <dbl> 10612.7, 11428.4, 5463.0, 3261.5, 2121.7, 1…
$ `Fe [µg/g] (Leachate)` <dbl> 5935.0, 6814.3, 2465.7, 3936.8, 189.6, 7711…
$ `Ba [µg/g] (Residue)` <dbl> 171.0, 198.0, 251.0, 290.0, 315.0, 259.0, 3…
$ `Sr [µg/g] (Residue)` <dbl> 66.6, 71.3, 90.3, 99.8, 106.0, 90.7, 108.0,…
$ `Ca [µg/g] (Residue)` <dbl> 2315.5, 2369.6, 3007.7, 3447.9, 3713.2, 331…
$ `Al [µg/g] (Residue)` <dbl> 29262.5, 35561.4, 45862.4, 52485.6, 55083.0…
$ `Fe [µg/g] (Residue)` <dbl> 14834.8, 18301.4, 24534.5, 30745.3, 28532.2…Clean the column names with Janitor
we can remove capitalization, spaces, and strange characters from the column names with the function clean_names() from the Janitor Package.
Rows: 128
Columns: 19
$ event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL73BC…
$ latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617, 39…
$ longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117, 24…
$ elevation_m <dbl> -339, -339, -339, -339, -339, -339, -339, -339,…
$ sample_label_barite_sr <chr> "SL73-1", "SL73-2", "SL73-3", "SL73-4", "SL73-5…
$ samp_type <chr> "non-S1", "non-S1", "non-S1", "S1b", "S1b", "S1…
$ depth_m <dbl> 0.0045, 0.0465, 0.0665, 0.1215, 0.1765, 0.1915,…
$ age_ka_bp <dbl> 1.66, 3.13, 4.13, 5.75, 7.30, 7.78, 8.65, 9.94,…
$ ca_co3_percent <dbl> 61.5, 55.1, 53.0, 43.4, 41.8, 42.3, 42.6, 39.1,…
$ ba_mg_g_leachate <dbl> 72.6, 64.3, 37.4, 63.5, 101.0, 141.0, 75.2, 99.…
$ sr_mg_g_leachate <dbl> 767, 681, 690, 552, 527, 528, 551, 482, 391, 70…
$ ca_mg_g_leachate <dbl> 188951.9, 163260.4, 162188.7, 125937.6, 124733.…
$ al_mg_g_leachate <dbl> 10612.7, 11428.4, 5463.0, 3261.5, 2121.7, 12740…
$ fe_mg_g_leachate <dbl> 5935.0, 6814.3, 2465.7, 3936.8, 189.6, 7711.3, …
$ ba_mg_g_residue <dbl> 171.0, 198.0, 251.0, 290.0, 315.0, 259.0, 310.0…
$ sr_mg_g_residue <dbl> 66.6, 71.3, 90.3, 99.8, 106.0, 90.7, 108.0, 96.…
$ ca_mg_g_residue <dbl> 2315.5, 2369.6, 3007.7, 3447.9, 3713.2, 3316.7,…
$ al_mg_g_residue <dbl> 29262.5, 35561.4, 45862.4, 52485.6, 55083.0, 44…
$ fe_mg_g_residue <dbl> 14834.8, 18301.4, 24534.5, 30745.3, 28532.2, 22…Watch out: Janitor transforms µ into m (so micrograms become milligrams).
Place fixed variables in the first columns
Which column is a fixed variable?
I’m not sure if ca_co3_percent is a measured variable, and if it belongs to another informational unit.
Besides that, the fixed variables are already in front.
There are values stored in the column names
Let’s pivot the measured variables.
pangaea_long <-
pangaea_data %>%
pivot_longer(
cols = contains(match = c('leachate', 'residue')),
values_to = 'concentration',
names_to = 'element'
)
pangaea_long# A tibble: 1,280 × 11
event latit…¹ longi…² eleva…³ sampl…⁴ samp_…⁵ depth_m age_k…⁶ ca_co…⁷ element
<chr> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <chr>
1 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ba_mg_…
2 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 sr_mg_…
3 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ca_mg_…
4 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 al_mg_…
5 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 fe_mg_…
6 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ba_mg_…
7 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 sr_mg_…
8 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ca_mg_…
9 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 al_mg_…
10 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 fe_mg_…
# … with 1,270 more rows, 1 more variable: concentration <dbl>, and abbreviated
# variable names ¹latitude, ²longitude, ³elevation_m,
# ⁴sample_label_barite_sr, ⁵samp_type, ⁶age_ka_bp, ⁷ca_co3_percent
# ℹ Use `print(n = ...)` to see more rows, and `colnames()` to see all variable namesRows: 1,280
Columns: 11
$ event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL73BC…
$ latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617, 39…
$ longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117, 24…
$ elevation_m <dbl> -339, -339, -339, -339, -339, -339, -339, -339,…
$ sample_label_barite_sr <chr> "SL73-1", "SL73-1", "SL73-1", "SL73-1", "SL73-1…
$ samp_type <chr> "non-S1", "non-S1", "non-S1", "non-S1", "non-S1…
$ depth_m <dbl> 0.0045, 0.0045, 0.0045, 0.0045, 0.0045, 0.0045,…
$ age_ka_bp <dbl> 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66,…
$ ca_co3_percent <dbl> 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5,…
$ element <chr> "ba_mg_g_leachate", "sr_mg_g_leachate", "ca_mg_…
$ concentration <dbl> 72.6, 767.0, 188951.9, 10612.7, 5935.0, 171.0, …When we pivot data we move them from a wide to a long format and vice-versa.
Pivot Longer
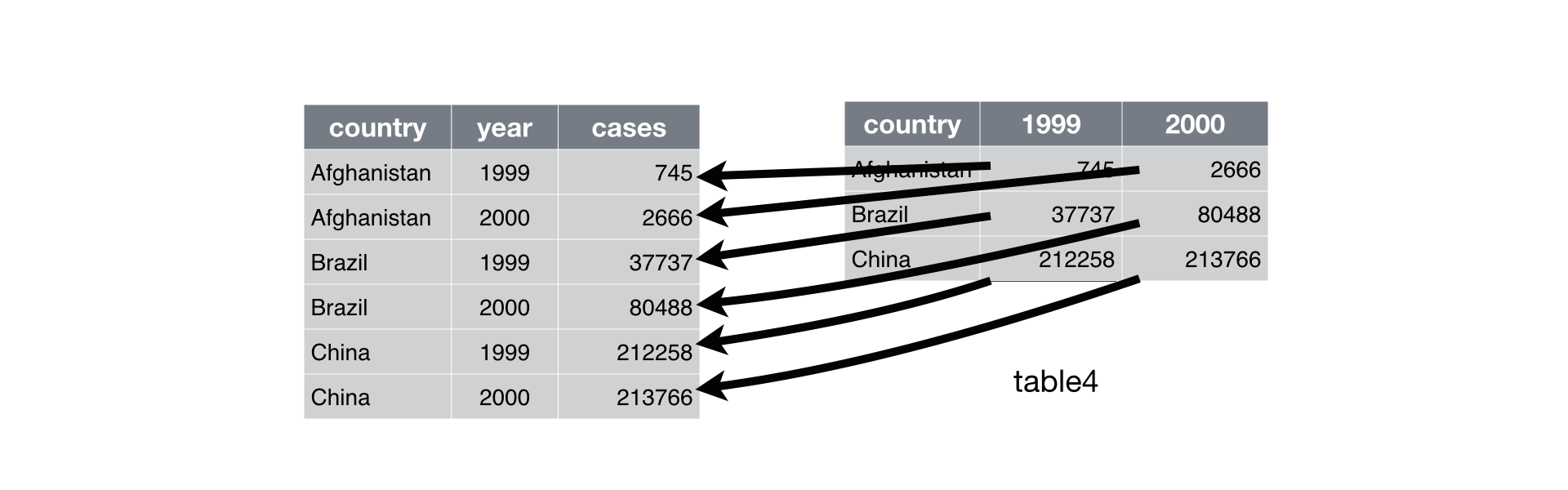
Source : R4DS - Tidy Data
Pivot Longer
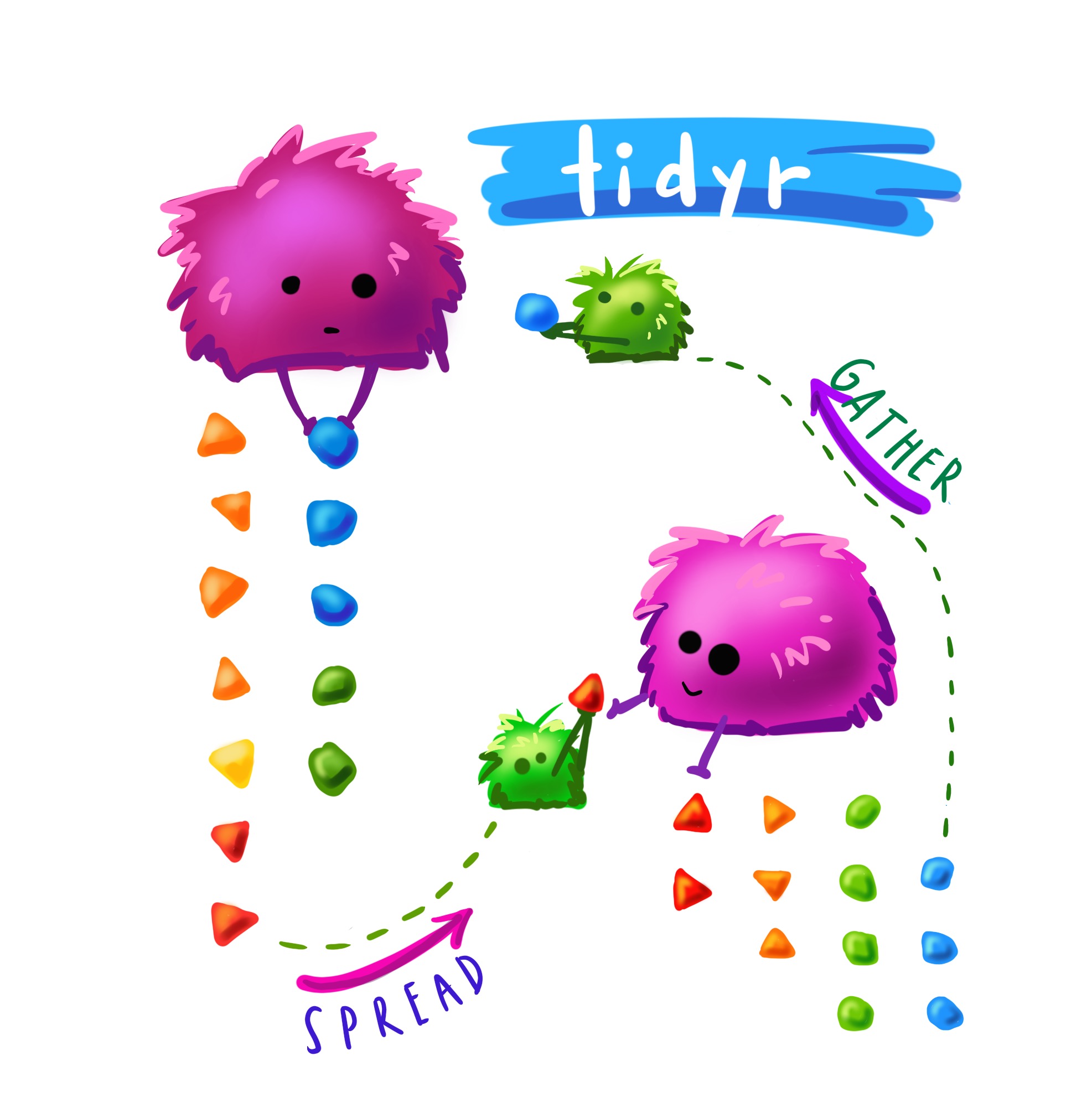
(Image from Alisson Horst, check her other stats illustrations here)
One cell contains multiple values
Now it’s clear that element contains more than one value.
For example: ba_mg_g_leachate is not a single values and could be split into:
- element:
ba. - unit:
mg_g. - fraction:
leachate.
Let’s split this column at the _ and reconstitute it in a tidy way
pangaea_tidy <-
pangaea_long %>%
separate(element, into = c('element', 'unit_num', 'unit_dem', 'fraction'), sep = '_') %>%
unite(col = 'unit', unit_num:unit_dem, sep = '/')
pangaea_tidy# A tibble: 1,280 × 13
event latit…¹ longi…² eleva…³ sampl…⁴ samp_…⁵ depth_m age_k…⁶ ca_co…⁷ element
<chr> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <chr>
1 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ba
2 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 sr
3 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ca
4 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 al
5 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 fe
6 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ba
7 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 sr
8 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ca
9 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 al
10 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 fe
# … with 1,270 more rows, 3 more variables: unit <chr>, fraction <chr>,
# concentration <dbl>, and abbreviated variable names ¹latitude, ²longitude,
# ³elevation_m, ⁴sample_label_barite_sr, ⁵samp_type, ⁶age_ka_bp,
# ⁷ca_co3_percent
# ℹ Use `print(n = ...)` to see more rows, and `colnames()` to see all variable namesRows: 1,280
Columns: 13
$ event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL73BC…
$ latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617, 39…
$ longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117, 24…
$ elevation_m <dbl> -339, -339, -339, -339, -339, -339, -339, -339,…
$ sample_label_barite_sr <chr> "SL73-1", "SL73-1", "SL73-1", "SL73-1", "SL73-1…
$ samp_type <chr> "non-S1", "non-S1", "non-S1", "non-S1", "non-S1…
$ depth_m <dbl> 0.0045, 0.0045, 0.0045, 0.0045, 0.0045, 0.0045,…
$ age_ka_bp <dbl> 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66,…
$ ca_co3_percent <dbl> 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5,…
$ element <chr> "ba", "sr", "ca", "al", "fe", "ba", "sr", "ca",…
$ unit <chr> "mg/g", "mg/g", "mg/g", "mg/g", "mg/g", "mg/g",…
$ fraction <chr> "leachate", "leachate", "leachate", "leachate",…
$ concentration <dbl> 72.6, 767.0, 188951.9, 10612.7, 5935.0, 171.0, …And let’s fix the measurement unit
Remember that janitor transformed µ into m?
# A tibble: 1,280 × 13
event latit…¹ longi…² eleva…³ sampl…⁴ samp_…⁵ depth_m age_k…⁶ ca_co…⁷ element
<chr> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <chr>
1 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ba
2 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 sr
3 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ca
4 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 al
5 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 fe
6 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ba
7 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 sr
8 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 ca
9 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 al
10 SL73… 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 fe
# … with 1,270 more rows, 3 more variables: unit <chr>, fraction <chr>,
# concentration <dbl>, and abbreviated variable names ¹latitude, ²longitude,
# ³elevation_m, ⁴sample_label_barite_sr, ⁵samp_type, ⁶age_ka_bp,
# ⁷ca_co3_percent
# ℹ Use `print(n = ...)` to see more rows, and `colnames()` to see all variable namesData can take many shapes
pangaea_also_tidy <-
pangaea_tidy %>%
pivot_wider(names_from = 'element', values_from = 'fraction')
pangaea_also_tidy # A tibble: 1,280 × 16
event latitude longi…¹ eleva…² sampl…³ samp_…⁴ depth_m age_k…⁵ ca_co…⁶ unit
<chr> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <chr>
1 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
2 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
3 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
4 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
5 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
6 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
7 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
8 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
9 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
10 SL73BC 39.7 24.5 -339 SL73-1 non-S1 0.0045 1.66 61.5 mg/g
# … with 1,270 more rows, 6 more variables: concentration <dbl>, ba <chr>,
# sr <chr>, ca <chr>, al <chr>, fe <chr>, and abbreviated variable names
# ¹longitude, ²elevation_m, ³sample_label_barite_sr, ⁴samp_type, ⁵age_ka_bp,
# ⁶ca_co3_percent
# ℹ Use `print(n = ...)` to see more rows, and `colnames()` to see all variable namesRows: 1,280
Columns: 16
$ event <chr> "SL73BC", "SL73BC", "SL73BC", "SL73BC", "SL73BC…
$ latitude <dbl> 39.6617, 39.6617, 39.6617, 39.6617, 39.6617, 39…
$ longitude <dbl> 24.5117, 24.5117, 24.5117, 24.5117, 24.5117, 24…
$ elevation_m <dbl> -339, -339, -339, -339, -339, -339, -339, -339,…
$ sample_label_barite_sr <chr> "SL73-1", "SL73-1", "SL73-1", "SL73-1", "SL73-1…
$ samp_type <chr> "non-S1", "non-S1", "non-S1", "non-S1", "non-S1…
$ depth_m <dbl> 0.0045, 0.0045, 0.0045, 0.0045, 0.0045, 0.0045,…
$ age_ka_bp <dbl> 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66, 1.66,…
$ ca_co3_percent <dbl> 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5, 61.5,…
$ unit <chr> "mg/g", "mg/g", "mg/g", "mg/g", "mg/g", "mg/g",…
$ concentration <dbl> 72.6, 767.0, 188951.9, 10612.7, 5935.0, 171.0, …
$ ba <chr> "leachate", NA, NA, NA, NA, "residue", NA, NA, …
$ sr <chr> NA, "leachate", NA, NA, NA, NA, "residue", NA, …
$ ca <chr> NA, NA, "leachate", NA, NA, NA, NA, "residue", …
$ al <chr> NA, NA, NA, "leachate", NA, NA, NA, NA, "residu…
$ fe <chr> NA, NA, NA, NA, "leachate", NA, NA, NA, NA, "re…Pivot Wider
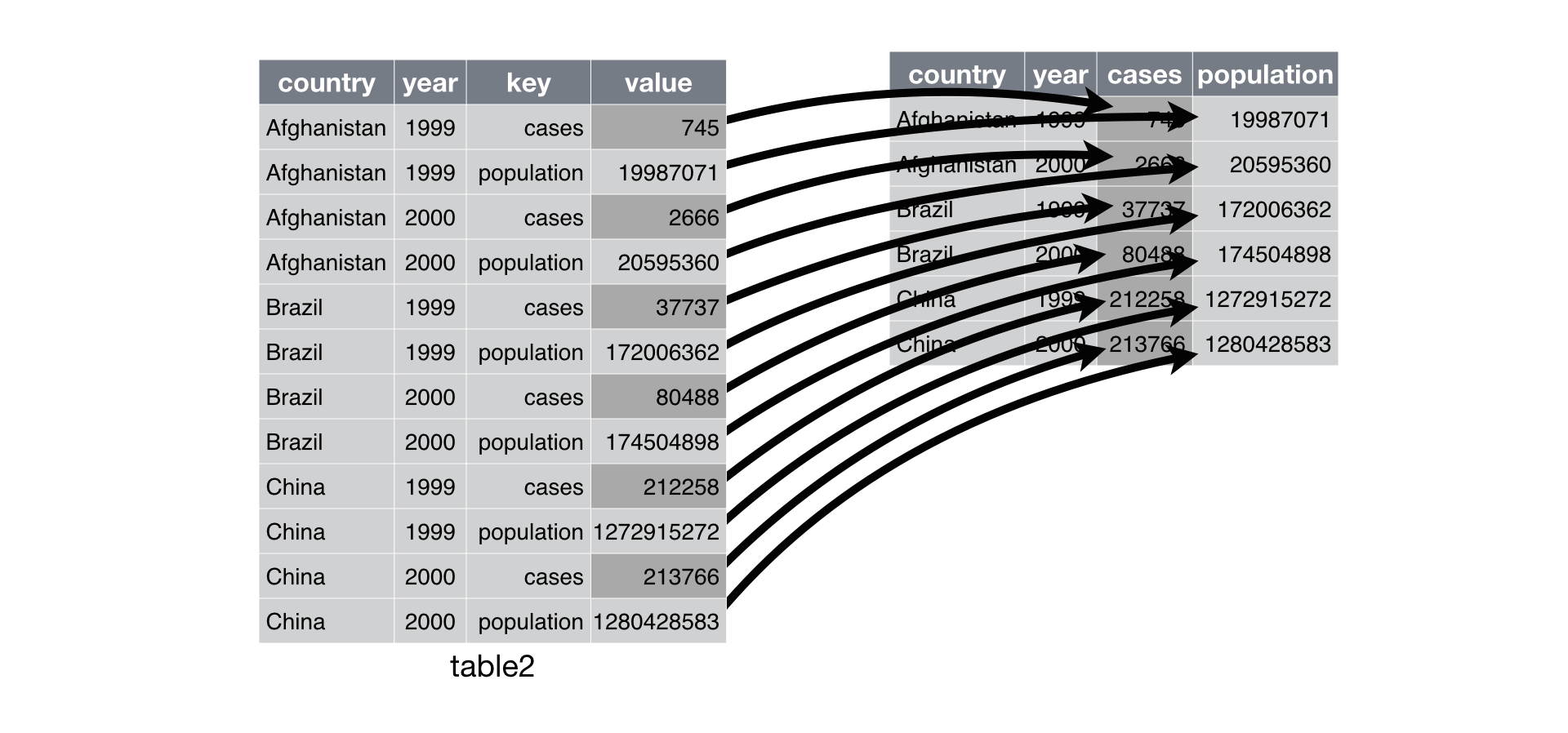
Source : R4DS - Tidy Data
Exercise
Tidy last week’s schedule.
